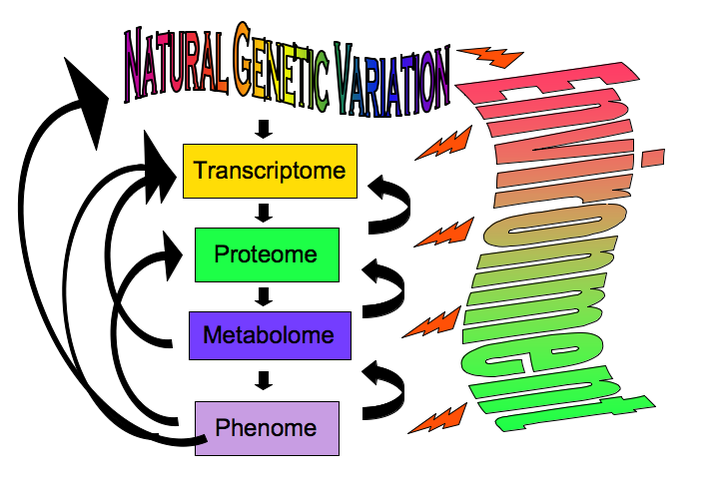
Mechanisms of Phenotypic VariationTo understand disease, ultimately we have to understand each omics level, how it evolves, and the mechanisms by which it can be perturbed. Phenotypic variation derives not only from genetic variation but also from the environment that can introduce both predictable and random perturbation of the physiological system. Metabolic homeostasis is achieved through interactions between different physiological or omics levels within an organism, and by feedback into the evolutionary genetics of the species through fitness effects.
|
A Drosophila Model for Metabolic Syndrome
Our primary current and future research plans use systems biology to correlate empirical data from across physiological levels such as RNA and metabolites, to decipher the relative contribution of natural genetic variation and environment to metabolic diseases like obesity and type-2 diabetes. Specifically, we are characterizing how variation in metabolic disease phenotypes maps to the metabolic pathway, by integrating metabolomic profiling with phenotypic, genomic, and gene expression data.
|
Metabolic Syndrome (MetS) is a constellation of symptoms such as obesity, elevated blood lipids, and insulin resistance that are predictive of type-2 diabetes and cardiovascular disease. MetS is a recently developed syndrome caused by our increased calorie intake and decreased exercise that exposes cryptic genetic variation: some people can eat high-fat, high-sugar diets without adverse impact while others are highly sensitive to their diet. Our research explores these effects using Drosophila, which also harbor cryptic genetic variation for disease, as a model to characterize the architecture of genotype-by-environment interactions of human-like metabolic disorders. With this charismatic model organism, we gain all the advantages of a carefully dissected metabolic and genetic system with substantial metabolic similarity to humans, while also being able to sample natural variation with high throughput genomic methods. Thus, using Drosophila, we can gain insights into a genetically complex disease that affects humans.
We have identified significant genotype-by-diet interactions for weight gain and other metabolic phenotypes in Drosophila, indicating that some genetic lines are metabolically sensitive to their diet while others are not. To identify metabolites that vary across genotypes and diets. Presently, funded by an NIH R01 we are genetically mapping QTL for both main effects and plastic (GxE) effects for metabolites and MetS related phenotypes. Future work will include characterizing genetic varation for exercise response and interaction of genetic effects with the gut microbiome. |
|
Evolutionary Ecology of Drosophila
Ecological context is essential to understanding the evolution of genetic architecture.
Our current work on Drosophila ecology is characterizing the species diversity and seasonal variation in species abundance of Drosophila in the vicinity of Tuscaloosa, AL. We are also exploring how mushroom eating fruit flies are able to detoxify the deadly alpha-amanitin toxin using metabolomic methods.
|